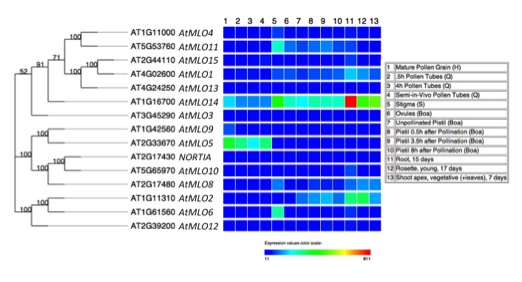
MLO genes and reproduction
NORTIA is a member of the MLO (Mildew Resistance Locus O) family of plant proteins that was originally discovered in the context of pathogen susceptibility in barley (Buschges et al., 1997). MLO proteins have seven predicted transmembrane domains and all members of the family have a calmodulin-binding domain in the C-terminal tail, suggesting that calcium signaling may be involved in MLO function (Devoto et al., 1999; Kim et al., 2002). In 2021, MLO proteins were discovered to have calcium channel activity when expressed in heterologous systems (Gao, et al., 2022). Wild-type versions of MLO genes are necessary for powdery mildew (PM) infection in both monocots and dicots, while mutations in these genes lead to resistance (Elliott et al., 2002; Consonni et al., 2006; Bai et al., 2008). Arabidopsis has fifteen MLO family members (Chen et al., 2006), but functions have been identified for only six. AtMLO2, AtMLO6, and AtMLO12 are involved in PM infection (Consonni et al., 2006), AtMLO4 and AtMLO11 are implicated in touch-induced root tropism (Chen et al., 2009), MLO1/5/9/15 are involved in pollen tube growth (Gao, et al. 2023), MLO15 is involved in root hair tip growth (Ogawa, et al. 2025), and NTA (AtMLO7) has a specific role in PT reception (Kessler et al., 2010). A common theme to these studies is that MLOs act downstream of receptor kinases to help cells respond to external stimuli.
Several of the Arabidopsis MLO genes are expressed in reproductive tissues. See our 2017 publication in Plant Reproduction for more information.
Selected MLO references
Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Topsch S, Vos P, Salamini F, Schulze-Lefert P (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695-705
Chen Z, Hartmann HA, Wu MJ, Friedman EJ, Chen JG, Pulley M, Schulze-Lefert P, Panstruga R, Jones AM (2006) Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol Biol 60: 583-597
Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, Somerville SC, Panstruga R (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38: 716-720
Davis TC, Jones DS, Dino AJ, Cejda NI, Yuan J, Willoughby AC, Kessler SA (2017) Arabidopsis thaliana MLO genes are expressed in discrete domains during reproductive development. Plant Reprod 30: 185-195
Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem 274: 34993-35004
Elliott C, Muller J, Miklis M, Bhat RA, Schulze-Lefert P, Panstruga R (2005) Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem J 385: 243-254
Gao Q, Wang C, Xi Y, Shao Q, Li L, Luan S. (2022) A receptor-channel trio conducts Ca2+ signalling for pollen tube reception. Nature. 607(7919):534-539.
Gao Q, Wang C, Xi Y, Shao Q, Hou C, Li L, Luan S. (2023) RALF signaling pathway activates MLO calcium channels to maintain pollen tube integrity. Cell Res. 33(1):71-79.
Jones DS, Kessler SA (2017) Cell type-dependent localization of MLO proteins. Plant Signal Behav 12: e1393135
Jones DS, Yuan J, Smith BE, Willoughby AC, Kumimoto EL, Kessler SA (2017) MILDEW RESISTANCE LOCUS O Function in Pollen Tube Reception Is Linked to Its Oligomerization and Subcellular Distribution. Plant Physiol 175: 172-185
Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968-971
Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447-451
Kusch S, Pesch L, Panstruga R (2016) Comprehensive Phylogenetic Analysis Sheds Light on the Diversity and Origin of the MLO Family of Integral Membrane Proteins. Genome Biol Evol 8: 878-895
Ogawa, ST, Zhang, W, Staiger, CS, and Kessler, SA. (2025). MLO-mediated Ca2+ influx regulates root hair tip growth in Arabidopsis. New Phytologist, in press.
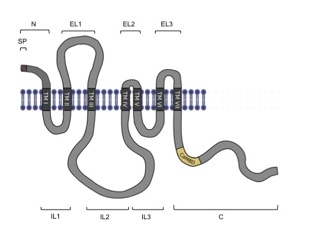
Predicted structure of MLO proteins
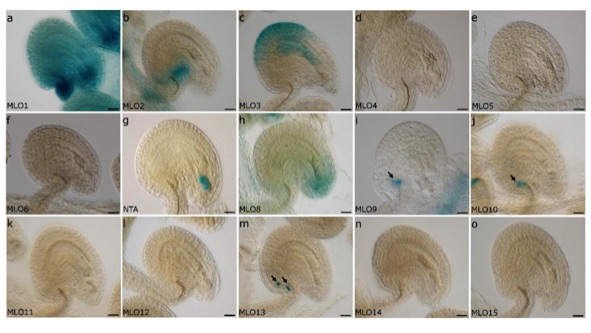
MLO expression in Arabidopsis ovules
Arabidopsis MLO phylogeny and gene expression comparison (generated by the Heat Tree Viewer)